Medical data is horrible to work with.
In medical imaging, data stores (archives) operate on clinical assumptions. Unfortunately, this means that when you want to extract an image (say a frontal chest x-ray), you will often get a folder full of other images with no easy way to tell them apart.

It makes sense that these are all the same folder, because in radiology we report cases, not images. This is everything the patient had scanned at the same time, after a traumatic injury.
Depending on the manufacturer, you might end up with horizontally or vertically flipped images. They might have inverted pixel values. They might be rotated. The question is, when dealing with a huge dataset (say, 50-100k images), how do you find these aberrations without having a doctor look at all of them?
You can try to code some elegant solution, like:
There are black borders at the sides of many chest x-rays (since most chests are taller than they are wide), so if there are more than 50 black pixel rows along the bottom, it is probably rotated 90 degrees
But as always, we run into failure modes.

Only the middle image here has the classic “black borders at the sides” appearance.
These brittle rules cannot solve these problems for us.
Enter software 2.0, where we use machine learning to build the solutions that we cannot put into code ourselves. Problems like rotated images are embarrassingly learnable. This means that, like humans, machines can very easily become (almost) perfect at these tasks.
So, the obvious answer is to use deep learning to fix our datasets for us. In this blog post I will show you where these techniques can work, how to do this with minimal effort, and present some examples of the methods in use. As an example, I will use the CXR14 dataset of Wang et al, which appears to have been pretty carefully curated but still contains the occasional bad image. I’ll even give you a new set of ~430 labels to exclude if you work with the CXR14 dataset, so you don’t have to worry about the bad images that snuck in!
So embarrassing
The first question we really need to ask is –
Q1: is the problem is embarrassingly learnable?
Considering most of the studies will be normal, you will need an enormously high precision to prevent excluding too many “good” studies. We should aim for something like 99.9%.
The cool thing is that problems this easy, where we can perform this well, are pretty visually identifiable. A good question to ask is “can you imagine a single visual rule to solve the problem?” This is certainly not the case when differentiating dogs from cats, which is the primary purpose of the ImageNet dataset :p
 There is too much variation, and too many similarities. I often use this example in talks – I can’t even imagine how to write rules that visually distinguish these two types of animal in a general sense. This is not embarrassingly learnable.
There is too much variation, and too many similarities. I often use this example in talks – I can’t even imagine how to write rules that visually distinguish these two types of animal in a general sense. This is not embarrassingly learnable.
But in medical data, many problems are really easy. There is very little variation. The anatomy, angles, lighting, distance, and background are all pretty stable. To show this, let’s look at a simple example from CXR14. Among the normal chest x-rays in the dataset, there are some that are rotated (and this is not identified in the labels, so we don’t know which ones). They can be rotated 90 degrees left or right, or 180 degrees (upside down).
Is this embarrassingly learnable?
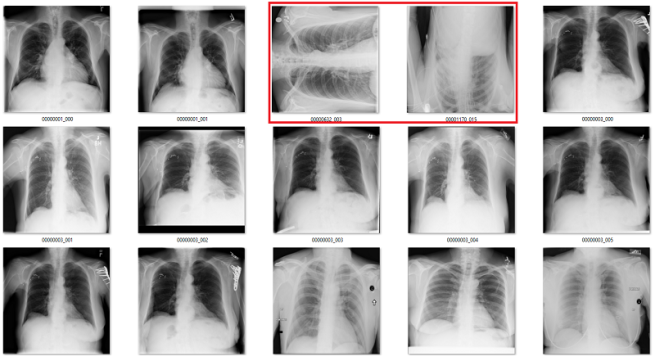
The difference between rotated and upright chest x-rays is really, embarrassingly simple.
The answer is yes. Visually, the abnormal studies are totally different from the normal studies. You could use a simple visual rule like “the shoulders should be above the heart” and you will get every example right. Given that anatomy is very stable, and all people have shoulders and hearts, this should be a learnable rule for a convolutional net.
So hungry
The second question we need to ask is –
Q2: do we have enough training data?
In the case of rotated images, of course we do; we can make it! All we need is a few thousand normal chest x-rays, and rotate them randomly. For example, if you are using numpy arrays for your images you might use a function like this:
def rotate(image):
rotated_image = np.rot90(image, k = np.random.choice(range(1,4)), axes = (1,2))
return rotated_image
This just rotates the image either 90, 180, or 270 degrees in the clockwise direction. In this case, rotating around the second and third axes, because the 1st axis is the number of channels (given theano dim ordering conventions).
Note: in this case, there are very few rotated images in the CXR14 dataset, so the chance of accidentally “correcting” an already rotated image is very small. We can just assume there are none in the data, and the model will learn well enough. If there were more abnormal images, then you might be better off hand-picking normal and abnormal ones. Since problems like rotation are so recognisable, I find I can label several thousand in an hour, so it doesn’t take a lot of effort. Since these problems are so simple, I often find I only need a few hundred examples to “solve” the challenge.
So we build a dataset of normal images, rotate half of them, and label them accordingly. In my case I selected 4000 training cases with 2000 of them rotated, and 2000 validation set cases with 1000 of them rotated. This seems like a nice amount of data (remember the rule of thumb: 1000 examples is probably good, plus a margin of error), and it fits in RAM so it is easy to train on my home computer.
For a fun change of pace in machine learning, I don’t need a separate test set. The proof is in the pudding – I’ll be running the model over the whole dataset anyway, and checking the outputs as my test.
In general for this sort of work I make life easy for myself. I shrink the images (since rotation detection doesn’t seem like it needs high resolution) to 256 x 256 pixels, and I use a pretrained resnet50 that comes with keras as the base network. There is no real reason to use a pretrained net, since almost any network you use will converge on a solution this simple, but it is easy and doesn’t slow anything down (training time is quick no matter what). I use a default set of parameters pretty much, and don’t do any tuning for a task this simple.
You can use whatever network you have on hand and coded. A VGG-net will work. A densenet will work. Anything will work, really.
After a few dozen epochs, I get my results (this is on the val set):
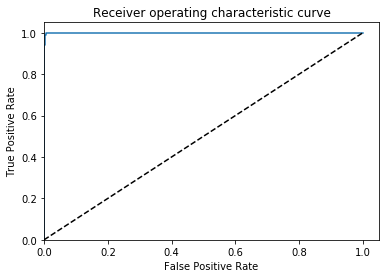
AUC = 0.999, ACC = 0.996, PREC = 0.998, REC = 0.994 🙂
Great, that is what I would hope to find if this is an embarrassingly learnable task.
Checking the results
As I have said previously, in medical image analysis we always need to check our results. Look at the images, make sure the model or process did what you wanted it to.
So the final step is to run the model across the entire dataset, make predictions, and then exclude the rotated studies*. Since there are almost no rotated studies in the data (and I know my recall is going to be very high), I can simply look at all the images that are predicted rotated.
If instead this was a problem with lots of abnormal images (say, more than 5% of the data) it would be more efficient to gather a few hundred random cases and hand-label a test set. Then you can keep track of how accurate your models are with the appropriate metrics.
I’m particularly concerned about any normal studies that got called rotated (false positives) because I don’t want to lose valuable training cases. This is actually a bigger concern than you might think, because the model is likely to over-call cases of a very specific type (maybe those that have the patient slouched and tilted), and if we exclude these as a rule we will introduce bias into our data and no longer have a “real-world” representative dataset. This obviously matters a great deal with medical data, since the whole goal is to produce systems that will work in real clinics.
The model identifies 171 cases as “rotated” in total. Interestingly, it actually functions as an “abnormal” detector, identifying a lot of bad cases that aren’t actually rotated. This makes sense, since it is probably learning anatomical landmarks. Anything abnormal, like rotated films or x-rays of other body parts, don’t have the same landmarks. So we actually get a broader yield than just finding the abnormally rotated images.
Of the 171 predictions, 51 are rotated frontal chest x-rays. Given the absurdly low prevalence (51 out of 120,000), this is already a fantastically low false positive rate.
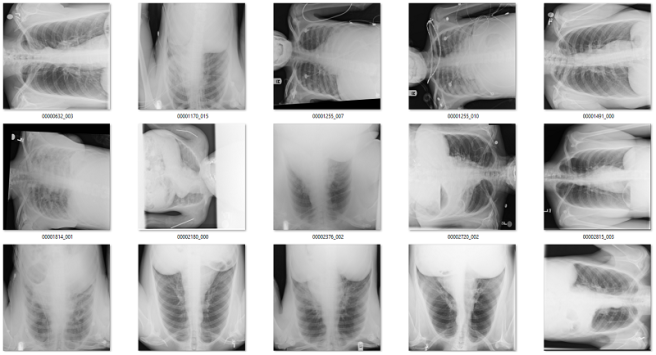
Examples of rotated chest films.
Of the remaining 120 cases, 56 are not frontal chest films. Mostly a mix of lateral films and abdominal x-rays. I would want to get rid of these anyway.

What about the rest? There are a mix of zoomed out studies (with big black or white borders), washed out studies (with the whole study grey), inverted pixel level studies, and so on.

Overall, there is about 10 studies that I would call clear false positives (meaning they are good frontal x-rays that I might want to save). Thankfully, even if you want to add them back in, with only 171 total predictions it is really easy to curate this by hand.
So the rotation detector looks like it partially solved some other issues (like pixel value inversions). To know how well it does, we will need to check if it missed other bad cases. We can test this, since pixel value inversions are easy to generate data for (for x in images, x = max – x).
So I trained a quick “inversion detector” with the same approach as the rotation detector, and ended up with great results.

AUC = 1.0, ACC = 0.9995, PREC = 0.999, REC= 1.0
Again, embarrassingly learnable. In this case there is probably some way to do this without machine learning (the histograms should look pretty different), but this is easy too.
So, did this specific detector find more inversions than the rotation detector did? Yep. The rotation detector found 4 in the whole dataset, and the inversion detector found 38 inverted studies. So the rotation detector finds some bad studies, but not all of them.
Take home point: Training individual models to solve each problem is the right way to go.
So, we need specific models for any additional cleaning tasks.
Every little bit helps
To show that a very small amount of labelled data can be useful, I took the lateral and bad regions films I found using the rotation detector (n = 56), and trained a new model on them. Since I didn’t have very many, I decided to go hog wild and didn’t even use a validation set. Since these tasks are embarrassingly learnable, once it gets near 100% it should generalise well. Obviously there is a risk of overtraining here, but I thought I would risk it.
It worked great! I found an extra couple of hundred lateral films, abdominal films, and a few pelvises.

Obviously if I was building this dataset from scratch it would be easier to solve this part, since I would have access to a lot of relevant non-frontal-chest images. For me to do better than I have now I would need to extract a set of images from a variety of body regions from my local hospital archive, and that is beyond the scope of this blog. So I can’t be sure I got most of these, but it is a pretty good effort from such a small dataset.
As an aside about the CXR14 data, one thing I noticed is that my models consistently freak out about films in young children. These paediatric films are different enough in appearance to to adult films that they are identified as “abnormal” by rotation detectors, inversion detectors, and bad-body-part detectors. I’d suggest that they should be ignored, but as the patient age is included in the label set so that can be done without deep learning involvement. Considering there are only 286 patients in the dataset under the age of 5, I would personally exclude all of them unless I was specifically trying to work on patients of that age group and really knew what I was doing, in a medical imaging sense. Really, I would probably exclude everyone under 10 years, since that is a reasonable age to consider the size and pathology profiles to be more “adult”. For interest, there are ~ 1400 cases under the age of 10, so about 1% of the data.
Take home point: paediatric chest x-rays are very different than adults. Considering only around 1% of the dataset is below the age of 10, they should probably be excluded unless you have a very good reason.
The badly positioned and zoomed films might be a problem, depending on your task, but arbitrarily defining a cut-off for what constitutes a “bad film” for all tasks isn’t something I want to do. Yet another thing that is task specific.
So that is about it. Overall, using deep learning to solve simple data cleaning problems works well. With about an hour of work, I have cleaned up the majority of the rotated and inverted images in the dataset (here is a csv of the filenames to watch out for). I have probably identified a fair chunk of the lateral films and films of other body parts, although to be sure I would need to build specific detectors for them. Without access to the original data, this would take too long for a simple blog post.
Looking at the CXR14 data more broadly, there weren’t a lot of image errors. The NIH team presumably curated their data fairly well. This is not always the case in medical datasets, and working out efficient ways to deal with the noise that comes from using clinical infrastructure for research tasks is super important if you want to build high performance medical AI systems.
Taking it further
So far, we have solved some very simple challenges, but not all of the problems we have in medical imaging are so easy.
Our team** applied these techniques when we were building a large hip fracture dataset. In particular, we excluded images from other body regions, we excluded cases with implanted metal (like hip replacements), and we zoomed in on the hip region to discard areas of the image that were unrelated to our question (hip fractures don’t occur outside the hip).
In the case of excluding metal, this was achieved with an automated text mining process – these prostheses are almost always reported when present, so I just found keywords that relate to implants. These labels were created in 10 minutes or so.
In the cases of wrong body part detection and bounding box prediction, there was no way to automatically generate the labels. So I made them myself. Even for something as complex as bounding box prediction (which is really an anatomical landmark identification task) we only needed about 750 cases. Each dataset only took an hour or so to make.
In this case, we used hand crafted test sets to quantify the results. From one of our papers:

Given that actually labelling the fractures took several months, an hour or two of extra labelling is a very small price to pay for a clean dataset. Particularly because the system can now take in any clinical image, and to the best of our knowledge can automatically exclude irrelevant or low quality films. This is exactly how a medical AI system needs to operate “in-the-wild”, unless you want to pay a human to hand-curate all of the images it analyses.
Summary
We all kind of accept that deep neural networks are about as good as humans at solving visual problems, given enough data. What is also obvious, but sometimes less spoken about, is that “enough data” is highly dependent on how hard the task is.
For a subset of medical image analysis problems, which happen to be the same problems we often want to solve when building medical datasets, the tasks are very easy and this makes the problems easy to solve with a small amount of data. Often, it takes less than an hour to identify image groups that would take a doctor hours upon hours to manually curate in each dataset.
As proof of the method and as thanks for reading my blog, I’ve provided a set of labels of ~430 bad images to exclude from the CXR14 dataset (as well as a recommendation that you exclude the ~1400 kids under the age of 10 unless you really know why you want to keep them). This won’t change any results for any papers, but the cleaner the images*** are for these datasets the better.
None of what I have demonstrated here is technically groundbreaking, which is why I’m not writing a formal paper about it. But for those of us who are building new datasets, particularly the doctors who don’t have a ton of experience with deep learning, I hope it might trigger some ideas about how software 2.0 could be able to solve your data problems with orders of magnitude less effort than a manual approach. The major barrier to building amazing medical AI systems at the moment is the huge cost of gathering and cleaning data, and in this context deep neural networks truly are unreasonably useful.
* BONUS NOTE TO EXPLAIN A BIT OF MY CASE REVIEW/LABELLING PROCESS
I check all my images in Windows file explorer! So low-brow 🙂

My actual workspace at the end of this blogpost, for the predictions of the rotation detector.
I simply transfer the cases that I want to see to a new folder, and then open the folder (with ‘extra-large icons’ as the view mode). The images at this size are about a quarter of your screen in height, and on most screens are large enough to detect big abnormalities like rotations. When I am labelling images with big abnormalities I simply ctrl-click all the examples in the folder, and then cut/paste them into a new folder. This is how I can do 1000/hr.
As janky as this system is, it is orders of magnitude better than most things I have tried from online repos or coded up myself.
The python code for moving the files around is really simple, but is some of my most used code when building data so I thought I would include it:
pos = rotation_labs[rotation_labs[‘Preds’] > 0.5]
#in this case, rotation_labs is a pandas dataframe which stores the image index/filename and the model prediction for that case. I subset it to have a dataframe of only the positive cases.
for i in pos[‘Index’]:
fname = “F:/cxr8/chest xrays/images/” + i
shutil.copy(fname, “F:/cxr8/data building/rotation/”)#all this does is copy the relevant images to a folder I have made called “rotation”.
Then I can go to that folder and have a look through. If I do a bit of manual curation and want to read the images back in, then it is a simple:
new_list = os.listdir(“F:/cxr8/data building/rotation/”)
** William Gale was my excellent co-author on this work. He managed to land an ML research position at Microsoft right out of his undergrad with us (!), and now focuses mostly on language problems. Keep an eye out for him, for sure 🙂
*** I know I promised to fix the labels a fair while ago, but I just got sidetracked with work for my PhD. I have actually been working on relabelling the data, so watch this space.


Cheers!
LikeLike
Lol! I check ‘em as thumbnails in explorer too!!!
LikeLike
Haha. 👍
LikeLiked by 1 person
Very cool post!
Looking forward to the relabeled CXR14 dataset. Properly labelled CXR14 would be of tremendous value to the community. Similar datasets in other tasks (like MINEX by NIST in fingerprint matching) allow to objectively judge progress in the field, something that medical imaging DL community badly lacks at the moment.
LikeLiked by 1 person
Yeah, I’m getting there. Unfortunately it isn’t embarrassingly buildable, but if it was then it wouldn’t be worth much anyway 🙂
LikeLiked by 2 people
Not like we could use the equivalent of a Grey-scale imagenet-like dataset trained over millions of epochs of augmentation on medical images…
LikeLiked by 1 person
Could you give me the datasets? Thank you very much. lsd201@126.com
LikeLiked by 1 person
Thanks for this blog.Could I translate your blog into Chinese? and publish it on my wordpress site,it a blog site ,too.I have learned machine learning for two years in the university.Look forward to your reply.
LikeLike
That is fine. Just link back to my blog 🙂
LikeLiked by 1 person
Of course I will.Have a nice day.
LikeLiked by 1 person
As a PACS manager in the NHS I have been constantly bemused at the way system vendors have failed to pick up on ML as a method of enabling some basic but productive housekeeping tasks. Body parts in the wrong exams; suspect laterality markers; inverted/rotated images, etc.
The focus appears to be on more complex problems such as identifying and classifying pathology, which is all very well but they are missing some really low hanging fruit in my opinion.
Perhaps you should offer some tips to the big players in this area?
LikeLiked by 1 person
@PACSMAN, if you think there is a way to bring such tools into the NHS without being a major PACS vendor, please drop me a line.
LikeLike
cool. indeed. could classification for medical images benefit from transfer learning (such as inception v3)? i have been trying without much success (compared to the non-transfer learning approach). regards
LikeLike