More than any other part of this blog series, what we talk about today will have the most impact on whether machines are going to replace doctors anytime soon.
We are going to start exploring the cutting edge of research in medical automation. In the previous articles in this series, we simply assumed deep learning can automate medical tasks. It made sense to do so, so we could get a bunch of concepts and definitions out of the way, but now we get to assess the claim directly.
One major focus of the coming pieces will be barriers to medical AI. So far we have discussed some external barriers to medical disruption, like regulation and the rate of automation, but we haven’t even touched on the technical challenges which could slow down the replacement of doctors.
Today we are going to explore a single research paper that is definitely state of the art. I wanted to review several articles in the one post, but each review is a large undertaking (this one is already 3000 words). There is just too much to talk about in each paper, so I will have to split this review of the evidence across several weeks. A blog series within a blog series.
For this week I offer many thanks to Dr Lily Peng, one of the authors of the paper, who thoroughly answered the methodological questions I had.
Since there is a lot to go over here, I will include another summary box at the end about my thoughts on this research, for the TL:DR crowd 🙂
Standard disclaimer: these posts are aimed at a broad audience including layfolk, machine learning experts, doctors and others. Experts will likely feel that my treatment of their discipline is fairly superficial, but will hopefully find a lot of interesting a new ideas outside of their domains. That said, if there are any errors please let me know so I can make corrections.
The state of the art
First up, I want to remind everyone – deep learning has really only been around as an applied method since 2012. So we haven’t even had five years to use this stuff in medicine, and us medical folks typically lag behind a bit. With that perspective some of these results are even more incredible, but we should acknowledge that this is just the beginning.
I’m going to review each paper I think is evidence of breakthrough medical automation, or that adds something useful to the conversation. I’ll describe the research, but spend time discussing a few key elements:
The task – is it a clinical task? How much of medical practice could be disrupted if it is automated? Why was this specific task chosen?
The data – how was the data collected and processed? How does it fit in to medical trials and regulatory requirements? What can we learn about the data needs of medical AI more broadly.
The results – do they equal or beat doctors? What exactly did they test? What more can we glean?
The conclusion – how big a deal is this? What does it show that we can extrapolate more broadly?
Google’s JAMA paper on diabetic retinopathy (December 2016)
The task:
Diabetic retinopathy is a major cause of blindness, caused by damage to the fine blood vessels in the back of the eye. This is diagnosed by looking at the back of the eye, where the vessels can be seen, a perceptual task.
A DL system might learn to recognise a white blotchy “cotton wool spots” pattern, for example.
They trained a deep learning system to perform several tasks related to diabetic retinopathy assessment. The headline result is the assessment of “referable” diabetic retinopathy, which is detecting moderate or worse eye disease (patients in this group are managed differently than those with “non-referable” eye disease). They also assessed the ability to identify severe retinopathy, and to detect macular oedema.
Data:
They trained the system with 130,000 retinal photos, each graded by 3 to 7 ophthalmologists with the final label decided by majority vote. The images were from a retrospective clinical dataset from 4 locations (EyePACS in the USA and 3 Indian hospitals), using a variety of cameras.
They validated the system on two datasets (in medicine, the term “validation” means patients that weren’t used for developing the system, synonymous with a test set in machine learning). One was a random sample of the EyePACS dataset and the other on a publicly available dataset from 3 French hospitals (Messidor-2). The latter dataset used a single camera for all pictures. These test sets were graded by a panel of 7-8 ophthalmologists, also using a majority vote mechanism.
The development/training data has a retinopathy prevalence of 55%, with 8% severe of worse. The validation data had far less disease, only 19.5% prevalence with only 1.7% severe or worse. This was intentional, the development set was enriched for positive cases (they added more than would normally occur in a clinical population).
Regarding the data quality, retinal photographs are typically between 1.3 and 3.5 megapixels in resolution. These images were shrunk to 299 pixels square, which is 0.08 megapixels (between 94% and 98% less pixels!). This is a baked-in property of the network architecture they applied, other image sizes cannot be used.
Network:
They used a pre-trained version of the Google Inception-v3 deep neural network, which is one of the best performing image analysis systems used today. Pre-training typically means they took a network already trained to detect non-medical objects (like photographs of cats and cars), and then trained it further on the specific medical images. This is why the network would only accept 229 x 299 pixel images.
Results:
This paper was what I consider the first major breakthrough in medical deep learning. They achieved equivalent performance to the individual ophthalmologists, as well as the “median” ophthalmologist, from their panel that they presented for comparison.
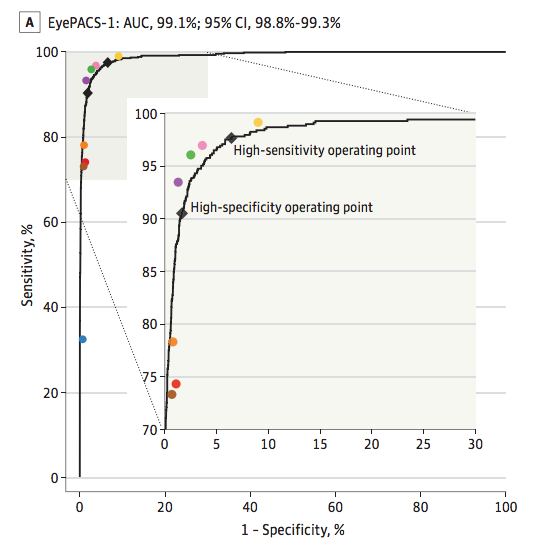
This is called an ROC curve, and is one of the best ways to judge diagnostic systems. The area under the curve (AUC) combines sensitivity and specificity in a single metric. 99.1% is very good.
The coloured dots are the ophthalmologists, and the black line is the deep learning system. As you can see, if you connect the coloured dots the actual ophthalmologists define a very similar ROC curve*. If you don’t understand ROC curves, you can just trust me that this is a valid way to show that the performance is about the same (and the FDA would agree with me).
They system was about as good at detecting macular oedema and a bit worse with severe retinopathy in absolute terms (AUC values), but head to head comparisons with the ophthalmologists were not published for these tasks.
Discussion:
There are a few interesting things to discuss about this research.
The cost: they paid a panel of ophthalmologists to label their data. I probably don’t need to explain how expensive a surgeon’s time is, right? Well, they got something like 500,000 labels in total. We must be talking millions of dollars, if they paid normal rates. That is more cash than most startups in this space would have on hand, and they certainly couldn’t afford to spend it on a single labeling task.
Statistically speaking, data is power. For medical AI, money makes data. ∴ Money is power, QED.
The task: they detected binary “referable eye disease” (moderate retinopathy or worse), severe retinopathy, and macular oedema from eye photographs. These are useful, clinically important tasks. Most importantly, these tasks cover the majority of what doctors do when they look in the eyes of patients with diabetes. Sure, this system will miss the occasional rare retinal melanoma, but for the day in/day out work of eye examination, this seems to be a pretty good analogue for human practice.
Data: the data was interesting for two reasons: quality and quantity.
We can see the quantity they needed, because they kindly ran some experiments. They tested how well their system performed with different numbers of training examples.
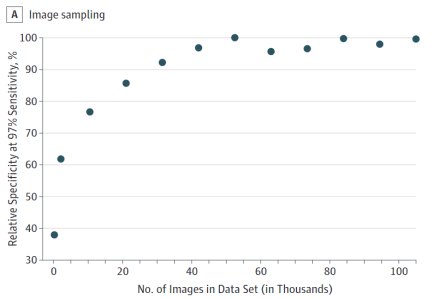
This shows us something pretty interesting. Their training (at least at the 97% sensitivity operating point) capped out at 60,000 cases. Notably, this one to two orders of magnitude larger than publicly available datasets, and I don’t doubt the need for data would increase if you went above 97% sensitivity.
The results tell us something else about the data size too. When they tried to replicate the work of ophthalmologists (the high specificity operating point), they got a sensitivity of 90% for referable disease but only 84% for severe or worse disease. It is possible that the task of identifying severe disease is simply harder, but I do note that there was 3 to 4 times as much training data for “moderate or worse” disease. There was less data in absolute terms (~9500 cases vs 34000 cases), but also in terms of prevalence (9% positive vs 30% positive).
Machine learning systems struggle with imbalanced data, and in my experience an imbalance worse than 30/70 is hard to deal with. It not only makes the training harder (less cases = less learning) but it makes the actual diagnosis harder (these systems develop a strong bias towards predicting the majority class).
We can see that they tried to deal with this problem. In screening populations the prevalence of “referable” disease is under 10%, so this is a highly imbalanced task. The team enriched their training set with additional positive cases, so the prevalence was about 30%. This seemed to work well, and the system performed strongly on the validation cohort (which had a clinical prevalence of around 8%). Bear in mind though that this method (enrichment of the minority class) only works when you have more positive cases, which is very rare. Other solutions to imbalanced data exist, but there is no real consensus on the best way to solve the problem.
There are two interesting things about the quality of the data.
Firstly, the downsampling/shrinking of the images. Isn’t is just ridiculous that this system can perform as well as human experts with 98% less pixels? We could say that since this system performs so well, most of those discarded pixels must be useless noise, which would otherwise make training deep learning systems harder. It is undoubtedly the case that humans are better at ignoring visual noise than computers, so maybe this is true.
But it may also be true that we have discarded useful information, and the system could perform even better on high resolution images. We can’t know because they couldn’t test it – the size reduction was required for this model to work.
The implication is actually broader though. Because deep learning systems have been largely developed to interpret small photographs, deep learning has never really been shown to work well in megapixel scale images**. It may actually be the case that higher resolution images are not usable, even if they contain more useful information.
So the down-sampling raises a few questions. Could deep learning have performed better with a higher resolution? Is low resolution fine for all medical tasks? Can we even use higher resolution images in deep learning from a technical perspective? We don’t know the answers for these questions, although we will try to narrow it down a bit as we look at some other papers in the coming weeks.
The second aspect of the data quality that is interesting is the quality of the labels. In machine learning, we need a good ground-truth. That means that we want the training data to be labeled correctly, so examples of retinopathy actually have retinopathy. This is harder said than done, because doctors disagree with each other. Again, the authors kindly provide the data.
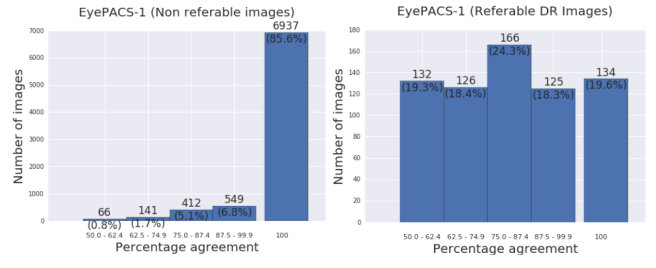
The panel of ophthalmologists agreed most of the time on non-referable disease, but disagreed most of the time when grading referable disease.
You can see that for moderate grade or above disease, at least one ophthalmologist had a different interpretation than the consensus opinion 80% of the time! This is why consensus labeling was used … it is meant to reduce the errors made by individuals.
Labeling errors will always exist in these sorts of datasets, and they will harm model performance. Deep learning can learn whatever you give it, it will learn to make incorrect diagnoses if you give it bad labels.
Consensus is not the only way around this problem. Some tasks have better ground truths, which we will see next week when I review a paper on dermatological lesions. Each lesion in that paper had a biopsy proven diagnosis, which is still interpreted by a pathologist but is much less variable. At the far extreme, some tasks have perfect labels. One of my own projects looks at a label which can’t be misinterpreted – mortality.
I think the key point about labels is that you get out what you put in. If you use labels from an individual doctor, at best you will be as good as that doctor. If you use consensus reporting, you should be able to marginally outperform individuals. And if you use perfectly defined ground-truths, you might be able to be perfect at the task.
Impact: Something like this could work very well as a screening tool, and I really appreciate the way the authors presented their results. They showed results for when the machine was operating like an ophthalmologist (lower false positive rate, but missing some positive cases) and they also showed results for when the system was optimised for screening (identifying almost all positive cases, but with a few more false positives).
This is an important thing to understand. These systems have a major advantage over human doctors: human doctors have a single operating point on a hypothetical ROC curve, a balance of sensitivity and specificity that is built on their experience and is very hard to change in any predictable way. In comparison, these systems can operate anywhere on their ROC curve without additional training. You can switch between diagnostic and screening modes with no additional cost, and know what the trade-offs will be without further experiments. This flexibility is super cool, and very useful when designing tests for actual clinical situations.
Thinking about regulation, this research is really close to being able to transition to clinical usage. They validated the model in a real world screening dataset, with multiple readers for each case. This is called an MRMC study (multireader, multicase), and is the general standard of evidence the FDA uses for computer assisted detection systems. Whether this holds with diagnostic systems isn’t entirely clear, but I wouldn’t be surprised if this system or one like it was FDA approved this year or next.
In terms of medical costs the impact of this task is moderate. Ophthalmology is not a large part of medicine, cost-wise, and looking in eyes is not a big ticket item in ophthalmology budgets.
The human impact could be huge though. There is an increasing epidemic of diabetes in the developing world and a massive shortage of eye specialists, so there is a compelling humanitarian role for this sort of technology. Considering the low resolution requirements of the processed images, if you paired this system with a low cost and easy to use handheld retinal camera it could improve millions of lives.
Ultimately though, even if retinopathy assessment is automated, the impact on medical jobs will be fairly limited. I actually think this as the sort of task which could easily result in increased demand for doctors when it is automated, as previously undiagnosed patients now need further assessment and treatment.
That is just in retinopathy assessment though. After we look at a few more papers, we will be able to think about some more general implications for the trajectory of medical automation.
So there you go. The first big breakthrough paper, from December 2016. Next week I will look at the paper from Stanford which claims their deep learning system can achieve “dermatologist-level classification of skin cancer”.
See you then 🙂
SUMMARY/TL:DR
- Google (and collaborators) trained a system to detect diabetic retinopathy (which causes 5% of blindness worldwide), and performed as well as a panel of ophthalmologists.
- This is a useful clinical task, which might not save a huge amount of money or displace doctors when automated but has a strong humanitarian motivation.
- They used 130,000 retinal images for training, 1 to 2 orders of magnitude larger than publicly available datasets.
- They enriched their training set with more positive cases, presumably to counteract the effects of training with imbalanced data (a problem with no consensus solution).
- The images were downsampled heavily, discarding >90% of the pixels, because most deep learning models are optimised for small photographs. We don’t know yet if this is a good thing to do.
- They used a panel of ophthalmologists to label the data, likely costing millions of dollars. This was done to achieve a “ground-truth” more accurate than the interpretation of any individual doctor.
- Points 5 and 6 are sources of error in all current medical deep learning systems, and this topic is poorly understood.
- Deep learning systems have an advantage over doctors in that they can be used at various “operating points”. The same system can perform high sensitivity screening and high specificity diagnosis without retraining. The trade-offs involved are transparent (unlike with doctors).
- This was an excellent piece of research. It is incredibly readable, and contains oodles of useful information in the text and the supplement.
- This research appears to match current FDA requirements for 510(k) approvals. While this technology is unlikely to go through that process, it is entirely possible this system or a derivative will be part of clinical practice in the next year or two.
*The distribution of ophthalmologists along the curve is pretty surprising to me, because it means that different doctors make very different predictions. Some of those doctors have 0 false positives, others have something like 10% false positives. It is a really wide range.
**Some work-arounds have been used, like cutting the images up into patches first. But this usually massively increases the number of negative examples, exacerbating the imbalanced data problem.
Posts in this series
Google and retinopathy
Next: Stanford and dermatology


About quality of labels, I remember a recent paper where they found benefit from explicitly modeling the labeling experts. In other words, instead of asking if this is a positive example, ask if expert 1 would call this a positive example, if expert 2 would call it, etc.
I found it with a little googling: “Who Said What: Modeling Individual Labelers Improves Classification” arxiv.org/abs/1703.08774
LikeLike
Interesting, I will check it out. Cheers.
LikeLike
Thanks for writing this series. One of the most interesting parts is the low res photos, I find it hard to understand that higher res photos give worse results. The data just seems missing in a low res photo.
Humans might take an overall view of certain features and then zoom in to have a closer look. Is human behavior of first identifying a feature and then zooming in closer to confirm the details something that has been tried with machine learning. Im not sure how it would be done.
LikeLike
The problem in high res photos is that you have increased the “dimensionality” of the problem. Each pixel is a new data point. So if, as in this example, 98% less pixels gives you the answer, adding all those pixels in is additional noise without any more signal.
Deep learning is very good at learning noise, so you are just muddying the answer.
Another way to think of it is that most convolutional filters look at between 3 and 5 pixel patches, with perhaps a field of view of 100 pixels at the final layer. If you have a megapixel image, you are only covering a tiny fraction of the total image, and there hasn’t been much work in architectures to adequately increase the field of view without massively increasing the size of the network.
LikeLike
Shouldn’t Google release the image dataset so that others can replicate or improve the results? It is absurd that Google can use its position in the market to do. There was and continues to be a fracas in the UK with Google/Deepmind using NHS patient data yet no one else has access to this data.
LikeLike
Well, they spent millions on it. Expecting them to give it away isn’t a very good way to promote research, particularly when no public institution has the capacity to do research on this scale.
The research community still benefits from the knowledge that comes out of the work, but any forced data sharing is just going to kill the whole field.
LikeLike
Millions of dollars is chicken feed for Google. Google makes tens of billions of dollars a year by placing ads between what consumers search for (free data collected by Google) and the result data (free data collected from websites).
The research community would benefit if Google gave data back. Since the data in this case is not huge, the research community would be able to work with it.
LikeLike
This was an interesting read. I am looking at applications of deep learning in medicine in India, so it was very relevant. Thank you! Keep doing them!
LikeLike
brilliant
LikeLike